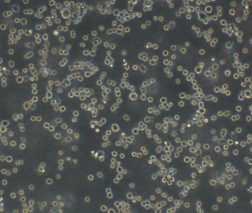
人多发性骨髓瘤细胞RPMI-8226
BLUEFBIO™ Product Sheet
|
细胞名称 |
人多发性骨髓瘤细胞RPMI-8226 |
|
|
|
货物编码 |
BFN608006107 |
||
|
产品规格 |
T25培养瓶x1 |
1.5ml冻存管x2 |
|
|
细胞数量 |
1x10^6 |
1x10^6 |
|
|
保存温度 |
37℃ |
-198℃ |
|
|
运输方式 |
常温保温运输 |
干冰运输 |
|
|
安全等级 |
1 |
||
|
用途限制 |
仅供科研用途 3类 |
||
|
培养体系 |
DMEM高糖培养基(Hyclone)+10%胎牛血清(Gibco)+1%双抗(Hyclone) |
||
|
培养温度 |
37℃ |
二氧化碳浓度 |
5% |
|
简介 |
人多发性骨髓瘤细胞RPMI-8226细胞来源于一位61岁的男性浆细胞瘤患者;可产生免疫球蛋白轻链,未检测到重链。 |
||
|
注释 |
Part of: Cancer Cell Line Encyclopedia (CCLE) project. Part of: COSMIC cell lines project. Part of: LL-100 blood cancer cell line panel. Part of: MD Anderson Cell Lines Project. Part of: NCI-60 cancer cell line panel. Part of: NCI-7 clinical proteomics reference material cell line panel. Characteristics: Produces IgG lambda. Doubling time: 60 hours (PubMed=25984343); 33.5 hours (NCI-DTP); ~60-70 hours (DSMZ). Microsatellite instability: Stable (MSS) (Sanger). Omics: Array-based CGH. Omics: CNV analysis. Omics: Deep antibody staining analysis. Omics: Deep exome analysis. Omics: Deep proteome analysis. Omics: Deep quantitative proteome analysis. Omics: Deep RNAseq analysis. Omics: DNA methylation analysis. Omics: Fluorescence phenotype profiling. Omics: lncRNA expression profiling. Omics: Metabolome analysis. Omics: Protein expression by reverse-phase protein arrays. Omics: SNP array analysis. Omics: Transcriptome analysis. Miscellaneous: HLA typing from personal communication of Pellat-Deceunynck C. Misspelling: RPMI-8266; Occasionally. Misspelling: RPMI-8826; In Cosmic 875867. Derived from sampling site: Peripheral blood. |
||
|
STR信息 |
Amelogenin:X,Y;CSF1PO:12;D13S317:11;D16S539:9;D18S51:15,19;D19S433:13.2,14;D21S11:28,29;D2S1338:20;D3S1358:16,17;D5S818:11,13;D7S820:9,10;D8S1179:12,13;FGA:19;TH01:8;TPOX:8,11;vWA:16,18; |
||
|
参考文献 |
PubMed=6042436; DOI=10.3181/00379727-125-32327 Matsuoka Y., Moore G.E., Yagi Y., Pressman D. Production of free light chains of immunoglobulin by a hematopoietic cell line derived from a patient with multiple myeloma. Proc. Soc. Exp. Biol. Med. 125:1246-1250(1967)
PubMed=4177524 Matsuoka Y., Takahashi M., Yagi Y., Moore G.E., Pressman D. Synthesis and secretion of immunoglobulins by established cell lines of human hematopoietic origin. J. Immunol. 101:1111-1120(1968)
PubMed=5243066 Moore G.E., Kitamura H. Cell line derived from patient with myeloma. N. Y. State J. Med. 68:2054-2060(1968)
PubMed=18605421; DOI=10.1093/jnci/45.5.951 Ikeuchi T., Sandberg A.A. Chromosome pulverization in virus-induced heterokaryons of mammalian cells from different species. J. Natl. Cancer Inst. 45:951-963(1970)
DOI=10.1016/B978-0-12-427150-0.50135-2 Matsuoka Y. Immunoglobulins. (In) Tissue culture. Methods and applications; Kruse P.F. Jr., Patterson M.K. Jr. (eds); pp.599-607, Academic Press; New York (1973)
PubMed=4366935 Minowada J., Nonoyama M., Moore G.E., Rauch A.M., Pagano J.S. The presence of the Epstein-Barr viral genome in human lymphoblastoid B-cell lines and its absence in a myeloma cell line. Cancer Res. 34:1898-1903(1974)
DOI=10.1007/978-1-4757-1647-4_11 Moore G.E. Cell lines from humans with hematopoietic malignancies. (In) Human tumor cells in vitro; Fogh J. (eds.); pp.299-331; Springer; New York (1975)
PubMed=3893568 Goldstein M., Hoxie J., Zembryki D., Matthews D., Levinson A.I. Phenotypic and functional analysis of B cell lines from patients with multiple myeloma. Blood 66:444-446(1985)
PubMed=2537114 Duperray C., Klein B., Durie B.G.M., Zhang X., Jourdan M., Poncelet P., Favier F., Vincent C., Brochier J., Lenoir G.M., Bataille R. Phenotypic analysis of human myeloma cell lines. Blood 73:566-572(1989)
PubMed=2140233; DOI=10.1111/j.1440-1827.1990.tb01549.x Nakano A., Harada T., Morikawa S., Kato Y. Expression of leukocyte common antigen (CD45) on various human leukemia/lymphoma cell lines. Acta Pathol. Jpn. 40:107-115(1990)
PubMed=2041050; DOI=10.1093/jnci/83.11.757 Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K., Vistica D.T., Hose C., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J. Natl. Cancer Inst. 83:757-766(1991)
CLPUB00447 Mulivor R.A., Suchy S.F. 1992/1993 catalog of cell lines. NIGMS human genetic mutant cell repository. 16th edition. October 1992. (In) Institute for Medical Research (Camden, N.J.) NIH 92-2011; pp.1-918; National Institutes of Health; Bethesda (1992)
PubMed=8943038; DOI=10.1073/pnas.93.24.13931 Bergsagel P.L., Chesi M., Nardini E., Brents L.A., Kirby S.L., Kuehl W.M. Promiscuous translocations into immunoglobulin heavy chain switch regions in multiple myeloma. Proc. Natl. Acad. Sci. U.S.A. 93:13931-13936(1996)
PubMed=9290701; DOI=10.1002/(SICI)1098-2744(199708)19:4<243::AID-MC5>3.0.CO;2-D Jia L.-Q., Osada M., Ishioka C., Gamo M., Ikawa S., Suzuki T., Shimodaira H., Niitani T., Kudo T., Akiyama M., Kimura N., Matsuo M., Mizusawa H., Tanaka N., Koyama H., Namba M., Kanamaru R., Kuroki T. Screening the p53 status of human cell lines using a yeast functional assay. Mol. Carcinog. 19:243-253(1997)
PubMed=9510473; DOI=10.1111/j.1349-7006.1998.tb00476.x Hosoya N., Hangaishi A., Ogawa S., Miyagawa K., Mitani K., Yazaki Y., Hirai H. Frameshift mutations of the hMSH6 gene in human leukemia cell lines. Jpn. J. Cancer Res. 89:33-39(1998)
PubMed=10087940; DOI=10.1016/S0165-4608(98)00157-5 Kuipers J., Vaandrager J.W., Weghuis D.O., Pearson P.L., Scheres J., Lokhorst H.M., Clevers H., Bast B.J.E.G. Fluorescence in situ hybridization analysis shows the frequent occurrence of 14q32.3 rearrangements with involvement of immunoglobulin switch regions in myeloma cell lines. Cancer Genet. Cytogenet. 109:99-107(1999)
PubMed=10557056; DOI=10.1038/sj.leu.2401563 Yoshida S., Nakazawa N., Iida S., Hayami Y., Sato S., Wakita A., Shimizu S., Taniwaki M., Ueda R. Detection of MUM1/IRF4-IgH fusion in multiple myeloma. Leukemia 13:1812-1816(1999)
PubMed=10583232; DOI=10.1046/j.1365-2141.1999.01705.x Puthier D., Derenne S., Barille S., Moreau P., Harousseau J.-L., Bataille R., Amiot M. Mcl-1 and Bcl-xL are co-regulated by IL-6 in human myeloma cells. Br. J. Haematol. 107:392-395(1999)
PubMed=10700174; DOI=10.1038/73432 Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V., Jeffrey S.S., van de Rijn M., Waltham M., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24:227-235(2000)
PubMed=10936422; DOI=10.1016/S0145-2126(99)00195-2 Drexler H.G., Matsuo Y. Malignant hematopoietic cell lines: in vitro models for the study of multiple myeloma and plasma cell leukemia. Leuk. Res. 24:681-703(2000)
DOI=10.1016/B978-0-12-221970-2.50457-5 Drexler H.G. The leukemia-lymphoma cell line factsbook. (In) ISBN 9780122219702; pp.1-733; Academic Press; London (2001)
PubMed=11154231; DOI=10.1182ood.v97.2.516 Hjertner O., Hjorth-Hansen H., Borset M., Seidel C., Waage A., Sundan A. Bone morphogenetic protein-4 inhibits proliferation and induces apoptosis of multiple myeloma cells. Blood 97:516-522(2001)
PubMed=11157491; DOI=10.1182ood.V97.3.729 Chesi M., Brents L.A., Ely S.A., Bais C., Robbiani D.F., Mesri E.A., Kuehl W.M., Bergsagel P.L. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood 97:729-736(2001)
PubMed=12068308; DOI=10.1038/nature00766 Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J.W., Woffendin H., Garnett M.J., Bottomley W., Davis N., Dicks E., Ewing R., Floyd Y., Gray K., Hall S., Hawes R., Hughes J., Kosmidou V., Menzies A., Mould C., Parker A., Stevens C., Watt S., Hooper S., Wilson R., Jayatilake H., Gusterson B.A., Cooper C., Shipley J.M., Hargrave D., Pritchard-Jones K., Maitland N.J., Chenevix-Trench G., Riggins G.J., Bigner D.D., Palmieri G., Cossu A., Flanagan A.M., Nicholson A., Ho J.W.C., Leung S.Y., Yuen S.T., Weber B.L., Seigler H.F., Darrow T.L., Paterson H., Marais R., Marshall C.J., Wooster R., Stratton M.R., Futreal P.A. Mutations of the BRAF gene in human cancer. Nature 417:949-954(2002)
PubMed=14555701 Shammas M.A., Shmookler Reis R.J., Akiyama M., Koley H., Chauhan D., Hideshima T., Goyal R.K., Hurley L.H., Anderson K.C., Munshi N.C. Telomerase inhibition and cell growth arrest by G-quadruplex interactive agent in multiple myeloma. Mol. Cancer Ther. 2:825-833(2003)
PubMed=15215163; DOI=10.1016/S0002-9440(10)63276-2 Inoue J., Otsuki T., Hirasawa A., Imoto I., Matsuo Y., Shimizu S., Taniwaki M., Inazawa J. Overexpression of PDZK1 within the 1q12-q22 amplicon is likely to be associated with drug-resistance phenotype in multiple myeloma. Am. J. Pathol. 165:71-81(2004)
PubMed=15748285; DOI=10.1186/1479-5876-3-11 Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. III, Stroncek D., Marincola F.M. HLA class I and II genotype of the NCI-60 cell lines. J. Transl. Med. 3:11-11(2005)
PubMed=16956823 Bataille R., Jego G., Robillard N., Barille-Nion S., Harousseau J.-L., Moreau P., Amiot M., Pellat-Deceunynck C. The phenotype of normal, reactive and malignant plasma cells Identification of 'many and multiple myelomas' and of new targets for myeloma therapy. Haematologica 91:1234-1240(2006)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433 Ikediobi O.N., Davies H., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Mironenko T., Parker A., Perry J., Raine K., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=17171682; DOI=10.1002/gcc.20404 Lombardi L., Poretti G., Mattioli M., Fabris S., Agnelli L., Bicciato S., Kwee I., Rinaldi A., Ronchetti D., Verdelli D., Lambertenghi-Deliliers G., Bertoni F., Neri A. Molecular characterization of human multiple myeloma cell lines by integrative genomics: insights into the biology of the disease. Genes Chromosomes Cancer 46:226-238(2007)
PubMed=18277095; DOI=10.4161/cbt.7.5.5712 Berglind H., Pawitan Y., Kato S., Ishioka C., Soussi T. Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination. Cancer Biol. Ther. 7:699-708(2008)
PubMed=18647998; DOI=10.1093/jncimonographs/lgn011 Dib A., Gabrea A., Glebov O.K., Bergsagel P.L., Kuehl W.M. Characterization of MYC translocations in multiple myeloma cell lines. J. Natl. Cancer Inst. Monogr. 39:25-31(2008)
PubMed=18700954; DOI=10.1186/1755-8794-1-37 Ronchetti D., Lionetti M., Mosca L., Agnelli L., Andronache A., Fabris S., Deliliers G.L., Neri A. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-342, and miR-561 with deregulated host genes in multiple myeloma. BMC Med. Genomics 1:37-37(2008)
PubMed=19306352; DOI=10.1002/gcc.20660 Lionetti M., Agnelli L., Mosca L., Fabris S., Andronache A., Todoerti K., Ronchetti D., Deliliers G.L., Neri A. Integrative high-resolution microarray analysis of human myeloma cell lines reveals deregulated miRNA expression associated with allelic imbalances and gene expression profiles. Genes Chromosomes Cancer 48:521-531(2009)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921 Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N. DNA fingerprinting of the NCI-60 cell line panel. Mol. Cancer Ther. 8:713-724(2009)
PubMed=20164919; DOI=10.1038/nature08768 Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F., Campbell P.J., Futreal P.A., Stratton M.R. Signatures of mutation and selection in the cancer genome. Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458 Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 70:2158-2164(2010)
PubMed=21173094; DOI=10.3324/haematol.2010.033456 Moreaux J., Klein B., Bataille R., Descamps G., Maiga S., Hose D., Goldschmidt H., Jauch A., Reme T., Jourdan M., Amiot M., Pellat-Deceunynck C. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica 96:574-582(2011)
PubMed=22384151; DOI=10.1371/journal.pone.0032096 Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T. Identification of cancer cell-line origins using fluorescence image-based phenomic screening. PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003 Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.K., Yu J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483:603-607(2012)
PubMed=22628656; DOI=10.1126/science.1218595 Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336:1040-1044(2012)
PubMed=23074853; DOI=10.1134/S1990519X12050136 Turilova V.K., Smirnova T.D. Karyotypic variability of human myeloma cell lines. Tsitologiia 54:621-636(2012)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342 Abaan O.D., Polley E.C., Davis S.R., Zhu Y.J., Bilke S., Walker R.L., Pineda M., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 73:4372-4382(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018 Moghaddas Gholami A., Hahne H., Wu Z., Auer F.J., Meng C., Wilhelm M., Kuster B. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 4:609-620(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20 Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A. The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer Metab. 1:20-20(2013)
PubMed=24670534; DOI=10.1371/journal.pone.0092047 Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C. High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner. PLoS ONE 9:E92047-E92047(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35 Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Hill Meyers B., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080 Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33:306-312(2015)
PubMed=25688540; DOI=10.1002to.a.22643 Maiga S., Brosseau C., Descamps G., Dousset C., Gomez-Bougie P., Chiron D., Menoret E., Kervoelen C., Vie H., Cesbron A., Moreau-Aubry A., Amiot M., Pellat-Deceunynck C. A simple flow cytometry-based barcode for routine authentication of multiple myeloma and mantle cell lymphoma cell lines. Cytometry A 87:285-288(2015)
PubMed=27377824; DOI=10.1038/sdata.2016.52 Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J. Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR. Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017 Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X., Egan R.K., Liu Q., Mironenko T., Mitropoulos X., Richardson L., Wang J., Zhang T., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J. A landscape of pharmacogenomic interactions in cancer. Cell 166:740-754(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4 Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H. A map of mobile DNA insertions in the NCI-60 human cancer cell panel. Mob. DNA 7:20-20(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005 Li J., Zhao W., Akbani R., Liu W., Ju Z., Ling S., Vellano C.P., Roebuck P., Yu Q., Eterovic A.K., Byers L.A., Davies M.A., Deng W., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y., Mills G.B., Liang H. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell 31:225-239(2017)
PubMed=29718670; DOI=10.1021/acs.jproteome.8b00165 Clark D.J., Hu Y., Bocik W., Chen L., Schnaubelt M., Roberts R., Shah P., Whiteley G., Zhang H. Evaluation of NCI-7 cell line panel as a reference material for clinical proteomics. J. Proteome Res. 17:2205-2215(2018)
PubMed=30285677; DOI=10.1186/s12885-018-4840-5 Tan K.-T., Ding L.-W., Sun Q.-Y., Lao Z.-T., Chien W., Ren X., Xiao J.-F., Loh X.-Y., Xu L., Lill M., Mayakonda A., Lin D.-C., Yang H., Koeffler H.P. Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines. BMC Cancer 18:940-940(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747 Dutil J., Chen Z., Monteiro A.N., Teer J.K., Eschrich S.A. An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines. Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3 Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J., Gelfand E.T., Bielski C.M., Li H., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569:503-508(2019)
PubMed=31160637; DOI=10.1038/s41598-019-44491-x Quentmeier H., Pommerenke C., Dirks W.G., Eberth S., Koeppel M., MacLeod R.A.F., Nagel S., Steube K., Uphoff C.C., Drexler H.G. The LL-100 panel: 100 cell lines for blood cancer studies. Sci. Rep. 9:8218-8218(2019)
PubMed=31465861; DOI=10.1016/j.jprot.2019.103504 Chanukuppa V., Paul D., Taunk K., Chatterjee T., Sharma S., Kumar S., Santra M.K., Rapole S. XPO1 is a critical player for bortezomib resistance in multiple myeloma: a quantitative proteomic approach. J. Proteomics 209:103504-103504(2019) |
||
验收细胞注意事项
1、收到人多发性骨髓瘤细胞RPMI-8226细胞,请查看瓶子是否有破裂,培养基是否漏出,是否浑浊,如有请尽快联系。
2、收到人多发性骨髓瘤细胞RPMI-8226细胞,如包装完好,请在显微镜下观察细胞。,由于运输过程中的问题,细胞培养瓶中的贴壁细胞有可能从瓶壁中脱落下来,显微镜下观察会出现细胞悬浮的情况,出现此状态时,请不要打开细胞培养瓶,应立即将培养瓶置于细胞培养箱里静止 3-5 小时左右,让细胞先稳定下,再于显微镜下观察,此时多数细胞会重新贴附于瓶壁。如细胞仍不能贴壁,请用台盼蓝染色法鉴定细胞活力,如台盼蓝染色证实细胞活力正常请按悬浮细胞的方法处理。
3、收到人多发性骨髓瘤细胞RPMI-8226细胞后,请镜下观察细胞,用恰当方式处理细胞。若悬浮的细胞较多,请离心收集细胞,接种到一个新的培养瓶中。弃掉原液,使用新鲜配制的培养基,使用进口胎牛血清。刚接到细胞,若细胞不多时 血清浓度可以加到 15%去培养。若细胞迏到 80%左右 ,血清浓度还是在 10%。
4、收到人多发性骨髓瘤细胞RPMI-8226细胞时如无异常情况 ,请在显微镜下观察细胞密度,如为贴壁细胞,未超过80%汇合度时,将培养瓶中培养基吸出,留下 5-10ML 培养基继续培养:超过 80%汇合度时,请按细胞培养条件传代培养。如为悬浮细胞,吸出培养液,1000 转/分钟离心 3 分钟,吸出上清,管底细胞用新鲜培养基悬浮细胞后移回培养瓶。
5、将培养瓶置于 37℃培养箱中培养,盖子微微拧松。吸出的培养基可以保存在灭菌过的瓶子里,存放于 4℃冰箱,以备不时之需。
6、24 小时后,人多发性骨髓瘤细胞RPMI-8226细胞形态已恢复并贴满瓶壁,即可传代。(贴壁细胞)将培养瓶里的培养基倒去,加 3-5ml(以能覆盖细胞生长面为准)PBS 或 Hanks’液洗涤后弃去。加 0.5-1ml 0.25%含 EDTA 的胰酶消化,消化时间以具体细胞为准,一般 1-3 分钟,不超过 5 分钟。可以放入37℃培养箱消化。轻轻晃动瓶壁,见细胞脱落下来,加入 3-5ml 培养基终止消化。用移液管轻轻吹打瓶壁上的细胞,使之完全脱落,然后将溶液吸入离心管内离心,1000rpm/5min。弃上清,视细胞数量决定分瓶数,一般一传二,如细胞量多可一传三,有些细胞不易传得过稀,有些生长较快的细胞则可以多传几瓶,以具体细胞和经验为准。(悬浮细胞)用移液管轻轻吹打瓶壁,直接将溶液吸入离心管离心即可。
7、贴壁细胞 ,悬浮细胞。严格无菌操作。换液时,换新的细胞培养瓶和换新鲜的培养液,37℃,5%CO2 培养。
特别提醒: 原瓶中培养基不宜继续使用,请更换新鲜培养基培养。