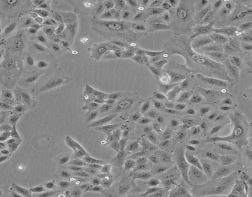
人肺癌细胞A549
BLUEFBIO™ Product Sheet
|
细胞名称 |
人肺癌细胞A549 |
|
|
|
货物编码 |
BFN60800665 |
||
|
产品规格 |
T25培养瓶x1 |
1.5ml冻存管x2 |
|
|
细胞数量 |
1x10^6 |
1x10^6 |
|
|
保存温度 |
37℃ |
-198℃ |
|
|
运输方式 |
常温保温运输 |
干冰运输 |
|
|
安全等级 |
1 |
||
|
用途限制 |
仅供科研用途 1类 |
||
|
培养体系 |
DMEM高糖培养基(Hyclone)+10%胎牛血清(Gibco)+1%双抗(Hyclone) |
||
|
培养温度 |
37℃ |
二氧化碳浓度 |
5% |
|
简介 |
人肺癌细胞A549细胞系是1972年由GiardDJ通过肺癌组织移植培养建系的,源自一位58岁的白人男性。A549能通过胞苷二磷脂酰胆碱途径合成富含不饱和脂肪酸的卵磷脂;角蛋白阳性。 |
||
|
注释 |
Part of: Cancer Cell Line Encyclopedia (CCLE) project. Part of: COSMIC cell lines project. Part of: ENCODE project common cell types; tier 2. Part of: JFCR39 cancer cell line panel. Part of: KuDOS 95 cell line panel. Part of: MD Anderson Cell Lines Project. Part of: Naval Biosciences Laboratory (NBL) collection (transferred to ATCC in 1982). Part of: NCI-60 cancer cell line panel. Part of: NCI-7 clinical proteomics reference material cell line panel. Doubling time: 18 hours (in RPMI 1640 + 10% FBS), 37 hours (in ACL-3), 36 hours (in ACL-3+BSA) (PubMed=3940644); 27.0 hours (PubMed=8286010); 22 hours (PubMed=25984343); 27 hours (from cell counting), 27 hours (from absorbance) (DOI=10.5897/IJBMBR2013.0154); 22.9 hours (NCI-DTP); ~28 hours (CLS); ~40 hours (DSMZ). Microsatellite instability: Stable (MSS) (PubMed=12661003; Sanger). Omics: Acetylation analysis by proteomics. Omics: Array-based CGH. Omics: CNV analysis. Omics: Deep antibody staining analysis. Omics: Deep exome analysis. Omics: Deep phosphoproteome analysis. Omics: Deep membrane proteome analysis. Omics: Deep proteome analysis. Omics: Deep RNAseq analysis. Omics: DNA methylation analysis. Omics: Fluorescence phenotype profiling. Omics: H3K4me3 ChIP-seq epigenome analysis. Omics: H3K9ac ChIP-seq epigenome analysis. Omics: lncRNA expression profiling. Omics: Metabolome analysis. Omics: Protein expression by reverse-phase protein arrays. Omics: Proteome analysis by 2D-DE/MS. Omics: shRNA library screening. Omics: SNP array analysis. Omics: Transcriptome analysis. Omics: Virome analysis using proteomics. Misspelling: A594; In PubMed=18227028. Misspelling: A59; In PubMed=16354588. |
||
|
STR信息 |
Amelogenin:X,Y;CSF1PO:10,12;D13S317:11;D16S539:11,12;D18S51:14,17;D19S433:13;D21S11:29;D2S1338:24;D3S1358:16;D5S818:11;D7S820:8,11;D8S1179:13,14;FGA:23;TH01:8,9.3;TPOX:8,11;vWA:14; |
||
|
参考文献 |
PubMed=4357758; DOI=10.1093/jnci/51.5.1417 Giard D.J., Aaronson S.A., Todaro G.J., Arnstein P., Kersey J.H., Dosik H., Parks W.P. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 51:1417-1423(1973)
PubMed=175022; DOI=10.1002/ijc.2910170110 Lieber M.M., Smith B.T., Szakal A., Nelson-Rees W.A., Todaro G.J. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int. J. Cancer 17:62-70(1976)
PubMed=327080; DOI=10.1093/jnci/59.1.221 Fogh J., Fogh J.M., Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J. Natl. Cancer Inst. 59:221-226(1977)
PubMed=833871; DOI=10.1093/jnci/58.2.209 Fogh J., Wright W.C., Loveless J.D. Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J. Natl. Cancer Inst. 58:209-214(1977)
PubMed=842942; DOI=10.1164/arrd.1977.115.2.285 Smith B.T. Cell line A549: a model system for the study of alveolar type II cell function. Am. Rev. Respir. Dis. 115:285-293(1977)
PubMed=924690; DOI=10.1002/ijc.2910200505 Kerbel R.S., Pross H.F., Leibovitz A. Analysis of established human carcinoma cell lines for lymphoreticular-associated membrane receptors. Int. J. Cancer 20:673-679(1977)
PubMed=22282976; DOI=10.1093/carcin/1.1.21 Day R.S. III, Ziolkowski C.H.J., Scudiero D.A., Meyer S.A., Mattern M.R. Human tumor cell strains defective in the repair of alkylation damage. Carcinogenesis 1:21-32(1980)
PubMed=6935474; DOI=10.1093/jnci/66.2.239 Wright W.C., Daniels W.P., Fogh J. Distinction of seventy-one cultured human tumor cell lines by polymorphic enzyme analysis. J. Natl. Cancer Inst. 66:239-247(1981)
PubMed=7459858 Rousset M., Zweibaum A., Fogh J. Presence of glycogen and growth-related variations in 58 cultured human tumor cell lines of various tissue origins. Cancer Res. 41:1165-1170(1981)
PubMed=7065527; DOI=10.1164/arrd.1982.125.2.222 Hay R.J., Williams C.D., Macy M.L., Lavappa K.S. Cultured cell lines for research on pulmonary physiology available through the American Type Culture Collection. Am. Rev. Respir. Dis. 125:222-232(1982)
PubMed=6148444; DOI=10.1093/jnci/73.4.801 Morstyn G., Russo A., Carney D.N., Karawya E., Wilson S.H., Mitchell J.B. Heterogeneity in the radiation survival curves and biochemical properties of human lung cancer cell lines. J. Natl. Cancer Inst. 73:801-807(1984)
PubMed=6500159; DOI=10.1159/000163283 Gershwin M.E., Lentz D., Owens R.B. Relationship between karyotype of tissue culture lines and tumorigenicity in nude mice. Exp. Cell Biol. 52:361-370(1984)
PubMed=3518877; DOI=10.3109/07357908609038260 Fogh J. Human tumor lines for cancer research. Cancer Invest. 4:157-184(1986)
PubMed=3940644 Brower M., Carney D.N., Oie H.K., Gazdar A.F., Minna J.D. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 46:798-806(1986)
PubMed=3945555; DOI=10.1093/nar/14.2.843 Valenzuela D.M., Groffen J. Four human carcinoma cell lines with novel mutations in position 12 of c-K-ras oncogene. Nucleic Acids Res. 14:843-852(1986)
PubMed=3129183 Hubbard W.C., Alley M.C., McLemore T.L., Boyd M.R. Evidence for thromboxane biosynthesis in established cell lines derived from human lung adenocarcinomas. Cancer Res. 48:2674-2677(1988)
PubMed=3335022 Alley M.C., Scudiero D.A., Monks A., Hursey M.L., Czerwinski M.J., Fine D.L., Abbott B.J., Mayo J.G., Shoemaker R.H., Boyd M.R. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 48:589-601(1988)
PubMed=3413074; DOI=10.1073/pnas.85.16.6042 Pereira-Smith O.M., Smith J.R. Genetic analysis of indefinite division in human cells: identification of four complementation groups. Proc. Natl. Acad. Sci. U.S.A. 85:6042-6046(1988)
PubMed=2388294; DOI=10.1093/jnci/82.17.1420 McLemore T.L., Litterst C.L., Coudert B.P., Liu M.C., Hubbard W.C., Adelberg S., Czerwinski M., McMahon N.A., Eggleston J.C., Boyd M.R. Metabolic activation of 4-ipomeanol in human lung, primary pulmonary carcinomas, and established human pulmonary carcinoma cell lines. J. Natl. Cancer Inst. 82:1420-1426(1990)
PubMed=2041050; DOI=10.1093/jnci/83.11.757 Monks A., Scudiero D.A., Skehan P., Shoemaker R.H., Paull K., Vistica D.T., Hose C., Langley J., Cronise P., Vaigro-Wolff A., Gray-Goodrich M., Campbell H., Mayo J.G., Boyd M.R. Feasibility of a high-flux anticancer drug screen using a diverse panel of cultured human tumor cell lines. J. Natl. Cancer Inst. 83:757-766(1991)
PubMed=8286010 Kiura K., Watarai S., Shibayama T., Ohnoshi T., Kimura I., Yasuda T. Inhibitory effects of cholera toxin on in vitro growth of human lung cancer cell lines. Anticancer Drug Des. 8:417-428(1993)
PubMed=7736387; DOI=10.1002/1097-0142(19950515)75:10<2442::AID-CNCR2820751009>3.0.CO;2-Q Campling B.G., Sarda I.R., Baer K.A., Pang S.C., Baker H.M., Lofters W.S., Flynn T.G. Secretion of atrial natriuretic peptide and vasopressin by small cell lung cancer. Cancer 75:2442-2451(1995)
PubMed=8626706; DOI=10.1074/jbc.271.19.11477 Quinn K.A., Treston A.M., Unsworth E.J., Miller M.-J., Vos M., Grimley C., Battey J., Mulshine J.L., Cuttitta F. Insulin-like growth factor expression in human cancer cell lines. J. Biol. Chem. 271:11477-11483(1996)
PubMed=9023415; DOI=10.1006/cimm.1996.1062 Seki N., Hoshino T., Kikuchi M., Hayashi A., Itoh K. HLA-A locus-restricted and tumor-specific CTLs in tumor-infiltrating lymphocytes of patients with non-small cell lung cancer. Cell. Immunol. 175:101-110(1997)
PubMed=9178645; DOI=10.1006/cimm.1997.1108 Nakao M., Sata M., Saitsu H., Yutani S., Kawamoto M., Kojiro M., Itoh K. CD4+ hepatic cancer-specific cytotoxic T lymphocytes in patients with hepatocellular carcinoma. Cell. Immunol. 177:176-181(1997)
PubMed=9290701; DOI=10.1002/(SICI)1098-2744(199708)19:4<243::AID-MC5>3.0.CO;2-D Jia L.-Q., Osada M., Ishioka C., Gamo M., Ikawa S., Suzuki T., Shimodaira H., Niitani T., Kudo T., Akiyama M., Kimura N., Matsuo M., Mizusawa H., Tanaka N., Koyama H., Namba M., Kanamaru R., Kuroki T. Screening the p53 status of human cell lines using a yeast functional assay. Mol. Carcinog. 19:243-253(1997)
PubMed=9636188; DOI=10.1073/pnas.95.13.7556 Kaplan D.H., Shankaran V., Dighe A.S., Stockert E., Aguet M., Old L.J., Schreiber R.D. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. U.S.A. 95:7556-7561(1998)
PubMed=9649128; DOI=10.1038/bjc.1998.361 Yi S., Chen J.-R., Viallet J., Schwall R.H., Nakamura T., Tsao M.-S. Paracrine effects of hepatocyte growth factor/scatter factor on non-small-cell lung carcinoma cell lines. Br. J. Cancer 77:2162-2170(1998)
PubMed=10358721; DOI=10.18926/AMO/31626 Matsushita A., Tabata M., Ueoka H., Kiura K., Shibayama T., Aoe K., Kohara H., Harada M. Establishment of a drug sensitivity panel using human lung cancer cell lines. Acta Med. Okayama 53:67-75(1999)
PubMed=10700174; DOI=10.1038/73432 Ross D.T., Scherf U., Eisen M.B., Perou C.M., Rees C., Spellman P.T., Iyer V., Jeffrey S.S., van de Rijn M., Waltham M., Pergamenschikov A., Lee J.C.F., Lashkari D., Shalon D., Myers T.G., Weinstein J.N., Botstein D., Brown P.O. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24:227-235(2000)
PubMed=11005564; DOI=10.1038/sj.neo.7900094 Kohno T., Sato T., Takakura S., Takei K., Inoue K., Nishioka M., Yokota J. Mutation and expression of the DCC gene in human lung cancer. Neoplasia 2:300-305(2000)
PubMed=11369051; DOI=10.1016/S0165-4608(00)00363-0 Luk C., Tsao M.-S., Bayani J., Shepherd F., Squire J.A. Molecular cytogenetic analysis of non-small cell lung carcinoma by spectral karyotyping and comparative genomic hybridization. Cancer Genet. Cytogenet. 125:87-99(2001)
PubMed=11958134 Zhan X.-Q., Guan Y.-J., Li C., Chen Z.-C., Xie J.-Y., Chen P., Liang S.-P. Differential proteomic analysis of human lung adenocarcinoma cell line A-549 and of normal cell line HBE. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao 34:50-56(2002)
PubMed=12661003; DOI=10.1002/gcc.10196 Seitz S., Wassmuth P., Plaschke J., Schackert H.K., Karsten U., Santibanez-Koref M.-F., Schlag P.M., Scherneck S. Identification of microsatellite instability and mismatch repair gene mutations in breast cancer cell lines. Genes Chromosomes Cancer 37:29-35(2003)
PubMed=12794755; DOI=10.1002/ijc.11184 Endoh H., Yatabe Y., Shimizu S., Tajima K., Kuwano H., Takahashi T., Mitsudomi T. RASSF1A gene inactivation in non-small cell lung cancer and its clinical implication. Int. J. Cancer 106:45-51(2003)
PubMed=14581340 Yokoi S., Yasui K., Iizasa T., Imoto I., Fujisawa T., Inazawa J. TERC identified as a probable target within the 3q26 amplicon that is detected frequently in non-small cell lung cancers. Clin. Cancer Res. 9:4705-4713(2003)
PubMed=15746151; DOI=10.1093/hmg/ddi092 Izumi H., Inoue J., Yokoi S., Hosoda H., Shibata T., Sunamori M., Hirohashi S., Inazawa J., Imoto I. Frequent silencing of DBC1 is by genetic or epigenetic mechanisms in non-small cell lung cancers. Hum. Mol. Genet. 14:997-1007(2005)
PubMed=15748285; DOI=10.1186/1479-5876-3-11 Adams S., Robbins F.-M., Chen D., Wagage D., Holbeck S.L., Morse H.C. III, Stroncek D., Marincola F.M. HLA class I and II genotype of the NCI-60 cell lines. J. Transl. Med. 3:11-11(2005)
PubMed=15900046; DOI=10.1093/jnci/dji133 Mashima T., Oh-hara T., Sato S., Mochizuki M., Sugimoto Y., Yamazaki K., Hamada J., Tada M., Moriuchi T., Ishikawa Y., Kato Y., Tomoda H., Yamori T., Tsuruo T. p53-defective tumors with a functional apoptosome-mediated pathway: a new therapeutic target. J. Natl. Cancer Inst. 97:765-777(2005)
PubMed=15999539 Matsumura T., Takigawa N., Kiura K., Shibayama T., Chikamori M., Tabata M., Ueoka H., Tanimoto M. Determinants of cisplatin and irinotecan activities in human lung adenocarcinoma cells: evidence of cisplatin accumulation and topoisomerase I activity. In Vivo 19:717-721(2005)
PubMed=17088437; DOI=10.1158/1535-7163.MCT-06-0433 Ikediobi O.N., Davies H., Bignell G.R., Edkins S., Stevens C., O'Meara S., Santarius T., Avis T., Barthorpe S., Brackenbury L., Buck G., Butler A., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Hunter C., Jenkinson A., Jones D., Kosmidou V., Lugg R., Menzies A., Mironenko T., Parker A., Perry J., Raine K., Richardson D., Shepherd R., Small A., Smith R., Solomon H., Stephens P., Teague J.W., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Reinhold W.C., Weinstein J.N., Stratton M.R., Futreal P.A., Wooster R. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol. Cancer Ther. 5:2606-2612(2006)
PubMed=17331233; DOI=10.1186/1471-2164-8-62 Nymark P., Lindholm P.M., Korpela M.V., Lahti L., Ruosaari S., Kaski S., Hollmen J., Anttila S., Kinnula V.L., Knuutila S. Gene expression profiles in asbestos-exposed epithelial and mesothelial lung cell lines. BMC Genomics 8:62-62(2007)
PubMed=17332333; DOI=10.1158/0008-5472.CAN-06-3339 Okabe T., Okamoto I., Tamura K., Terashima M., Yoshida T., Satoh T., Takada M., Fukuoka M., Nakagawa K. Differential constitutive activation of the epidermal growth factor receptor in non-small cell lung cancer cells bearing EGFR gene mutation and amplification. Cancer Res. 67:2046-2053(2007)
PubMed=17511773; DOI=10.1111/j.1349-7006.2007.00507.x Fukuyama T., Ichiki Y., Yamada S., Shigematsu Y., Baba T., Nagata Y., Mizukami M., Sugaya M., Takenoyama M., Hanagiri T., Sugio K., Yasumoto K. Cytokine production of lung cancer cell lines: correlation between their production and the inflammatory/immunological responses both in vivo and in vitro. Cancer Sci. 98:1048-1054(2007)
PubMed=18083107; DOI=10.1016/j.cell.2007.11.025 Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J.M., Yuan J., Bakalarski C.E., Villen J., Kornhauser J.M., Smith B., Li D., Zhou X., Gygi S.P., Gu T.-L., Polakiewicz R.D., Rush J., Comb M.J. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131:1190-1203(2007)
PubMed=19372543; DOI=10.1158/1535-7163.MCT-08-0921 Lorenzi P.L., Reinhold W.C., Varma S., Hutchinson A.A., Pommier Y., Chanock S.J., Weinstein J.N. DNA fingerprinting of the NCI-60 cell line panel. Mol. Cancer Ther. 8:713-724(2009)
PubMed=19472407; DOI=10.1002/humu.21028 Blanco R., Iwakawa R., Tang M., Kohno T., Angulo B., Pio R., Montuenga L.M., Minna J.D., Yokota J., Sanchez-Cespedes M. A gene-alteration profile of human lung cancer cell lines. Hum. Mutat. 30:1199-1206(2009)
PubMed=19608861; DOI=10.1126/science.1175371 Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834-840(2009)
PubMed=19657000 Rubporn A., Srisomsap C., Subhasitanont P., Chokchaichamnankit D., Chiablaem K., Svasti J., Sangvanich P. Comparative proteomic analysis of lung cancer cell line and lung fibroblast cell line. Cancer Genomics Proteomics 6:229-237(2009)
PubMed=20164919; DOI=10.1038/nature08768 Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C., Widaa S., Hinton J., Fahey C., Fu B., Swamy S., Dalgliesh G.L., Teh B.T., Deloukas P., Yang F., Campbell P.J., Futreal P.A., Stratton M.R. Signatures of mutation and selection in the cancer genome. Nature 463:893-898(2010)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458 Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 70:2158-2164(2010)
PubMed=20545000; DOI=10.3724/SP.J.1141.2010.02113 Peng K.-J., Wang J.-H., Su W.-T., Wang X.-C., Yang F.-T., Nie W.-H. Characterization of two human lung adenocarcinoma cell lines by reciprocal chromosome painting. Dong Wu Xue Yan Jiu 31:113-121(2010)
PubMed=21060948; DOI=10.1039/c0mb00055h Yu G., Xiao C.-L., Lu C.-H., Jia H.-T., Ge F., Wang W., Yin X.-F., Jia H.-L., He J.-X., He Q.-Y. Phosphoproteome profile of human lung cancer cell line A549. Mol. Biosyst. 7:472-479(2011)
PubMed=22278370; DOI=10.1074/mcp.M111.014050 Geiger T., Wehner A., Schaab C., Cox J., Mann M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics 11:M111.014050-M111.014050(2012)
PubMed=22313637; DOI=10.4161/cbt.19238 Takata M., Chikumi H., Miyake N., Adachi K., Kanamori Y., Yamasaki A., Igishi T., Burioka N., Nanba E., Shimizu E. Lack of AKT activation in lung cancer cells with EGFR mutation is a novel marker of cetuximab sensitivity. Cancer Biol. Ther. 13:369-378(2012)
PubMed=22336246; DOI=10.1016/j.bmc.2012.01.017 Kong D., Yamori T. JFCR39, a panel of 39 human cancer cell lines, and its application in the discovery and development of anticancer drugs. Bioorg. Med. Chem. 20:1947-1951(2012)
PubMed=22384151; DOI=10.1371/journal.pone.0032096 Lee J.-S., Kim Y.K., Kim H.J., Hajar S., Tan Y.L., Kang N.-Y., Ng S.H., Yoon C.N., Chang Y.-T. Identification of cancer cell-line origins using fluorescence image-based phenomic screening. PLoS ONE 7:E32096-E32096(2012)
PubMed=22460905; DOI=10.1038/nature11003 Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.K., Yu J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483:603-607(2012)
PubMed=22481983; DOI=10.2174/1874091X01206010016 Chauhan V., Howland M., Wilkins R. Effects of alpha-particle radiation on microRNA responses in human cell-lines. Open Biochem. J. 6:16-22(2012)
PubMed=22576795; DOI=10.3892/or.2012.1763 Ashinuma H., Takiguchi Y., Kitazono S., Kitazono-Saitoh M., Kitamura A., Chiba T., Tada Y., Kurosu K., Sakaida E., Sekine I., Tanabe N., Iwama A., Yokosuka O., Tatsumi K. Antiproliferative action of metformin in human lung cancer cell lines. Oncol. Rep. 28:8-14(2012)
PubMed=22628656; DOI=10.1126/science.1218595 Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336:1040-1044(2012)
PubMed=22961666; DOI=10.1158/2159-8290.CD-12-0112 Byers L.A., Wang J., Nilsson M.B., Fujimoto J., Saintigny P., Yordy J., Giri U., Peyton M., Fan Y.H., Diao L., Masrorpour F., Shen L., Liu W., Duchemann B., Tumula P., Bhardwaj V., Welsh J., Weber S., Glisson B.S., Kalhor N., Wistuba I.I., Girard L., Lippman S.M., Mills G.B., Coombes K.R., Weinstein J.N., Minna J.D., Heymach J.V. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2:798-811(2012)
DOI=10.5897/IJBMBR2013.0154 Iloki Assanga S.B., Gil-Salido A.A., Lewis Lujan L.M., Rosas-Durazo A., Acosta-Silva A.L., Rivera-Castaneda E.G., Rubio-Pino J.L. Cell growth curves for different cell lines and their relationship with biological activities. Int. J. Biotechnol. Mol. Biol. Res. 4:60-70(2013)
PubMed=23325432; DOI=10.1101/gr.147942.112 Varley K.E., Gertz J., Bowling K.M., Parker S.L., Reddy T.E., Pauli-Behn F., Cross M.K., Williams B.A., Stamatoyannopoulos J.A., Crawford G.E., Absher D.M., Wold B.J., Myers R.M. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23:555-567(2013)
PubMed=23404217; DOI=10.1007/s11626-013-9581-9 Chang D., Xu H.-W., Guo Y.-H., Jiang X., Liu Y., Li K.-L., Pan C.-X., Yuan M., Wang J.-F., Li T.-Z., Liu C.-T. Simulated microgravity alters the metastatic potential of a human lung adenocarcinoma cell line. In Vitro Cell. Dev. Biol. Anim. 49:170-177(2013)
PubMed=23856246; DOI=10.1158/0008-5472.CAN-12-3342 Abaan O.D., Polley E.C., Davis S.R., Zhu Y.J., Bilke S., Walker R.L., Pineda M., Gindin Y., Jiang Y., Reinhold W.C., Holbeck S.L., Simon R.M., Doroshow J.H., Pommier Y., Meltzer P.S. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 73:4372-4382(2013)
PubMed=23933261; DOI=10.1016/j.celrep.2013.07.018 Moghaddas Gholami A., Hahne H., Wu Z., Auer F.J., Meng C., Wilhelm M., Kuster B. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 4:609-620(2013)
PubMed=24135919; DOI=10.1038/ncomms3617 Balbin O.A., Prensner J.R., Sahu A., Yocum A., Shankar S., Malik R., Fermin D., Dhanasekaran S.M., Chandler B., Thomas D., Beer D.G., Cao X., Nesvizhskii A.I., Chinnaiyan A.M. Reconstructing targetable pathways in lung cancer by integrating diverse omics data. Nat. Commun. 4:2617-2617(2013)
PubMed=24279929; DOI=10.1186/2049-3002-1-20 Dolfi S.C., Chan L.L.-Y., Qiu J., Tedeschi P.M., Bertino J.R., Hirshfield K.M., Oltvai Z.N., Vazquez A. The metabolic demands of cancer cells are coupled to their size and protein synthesis rates. Cancer Metab. 1:20-20(2013)
PubMed=24618588; DOI=10.1371/journal.pone.0091433 Chernobrovkin A.L., Zubarev R.A. Detection of viral proteins in human cells lines by xeno-proteomics: elimination of the last valid excuse for not testing every cellular proteome dataset for viral proteins. PLoS ONE 9:E91433-E91433(2014)
PubMed=24670534; DOI=10.1371/journal.pone.0092047 Varma S., Pommier Y., Sunshine M., Weinstein J.N., Reinhold W.C. High resolution copy number variation data in the NCI-60 cancer cell lines from whole genome microarrays accessible through CellMiner. PLoS ONE 9:E92047-E92047(2014)
PubMed=25120651; DOI=10.3892/ol.2014.2234 Suzuki M., Ikeda K., Shiraishi K., Eguchi A., Mori T., Yoshimoto K., Shibata H., Ito T., Baba Y., Baba H. Aberrant methylation and silencing of IRF8 expression in non-small cell lung cancer. Oncol. Lett. 8:1025-1030(2014)
PubMed=25475639; DOI=10.5731ajpst.2014.01027 Shabram P., Kolman J.L. Evaluation of A549 as a new vaccine cell substrate: digging deeper with massively parallel sequencing. PDA J. Pharm. Sci. Technol. 68:639-650(2014)
PubMed=25960936; DOI=10.4161/21624011.2014.954893 Boegel S., Lower M., Bukur T., Sahin U., Castle J.C. A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines. OncoImmunology 3:E954893-E954893(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35 Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Hill Meyers B., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080 Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33:306-312(2015)
PubMed=26202522; DOI=10.1021/acs.jproteome.5b00477 Kitata R.B., Dimayacyac-Esleta B.R., Choong W.-K., Tsai C.-F., Lin T.-D., Tsou C.-C., Weng S.-H., Chen Y.-J., Yang P.-C., Arco S.D., Nesvizhskii A.I., Sung T.-Y., Chen Y.-J. Mining missing membrane proteins by high-pH reverse-phase stagetip fractionation and multiple reaction monitoring mass spectrometry. J. Proteome Res. 14:3658-3669(2015)
PubMed=26554430; DOI=10.1021/acs.analchem.5b03639 Dimayacyac-Esleta B.R., Tsai C.-F., Kitata R.B., Lin P.-Y., Choong W.-K., Lin T.-D., Wang Y.-T., Weng S.-H., Yang P.-C., Arco S.D., Sung T.-Y., Chen Y.-J. Rapid high-pH reverse phase stagetip for sensitive small-scale membrane proteomic profiling. Anal. Chem. 87:12016-12023(2015)
PubMed=27377824; DOI=10.1038/sdata.2016.52 Mestdagh P., Lefever S., Volders P.-J., Derveaux S., Hellemans J., Vandesompele J. Long non-coding RNA expression profiling in the NCI60 cancer cell line panel using high-throughput RT-qPCR. Sci. Data 3:160052-160052(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017 Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X., Egan R.K., Liu Q., Mironenko T., Mitropoulos X., Richardson L., Wang J., Zhang T., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J. A landscape of pharmacogenomic interactions in cancer. Cell 166:740-754(2016)
PubMed=27807467; DOI=10.1186/s13100-016-0078-4 Zampella J.G., Rodic N., Yang W.R., Huang C.R.L., Welch J., Gnanakkan V.P., Cornish T.C., Boeke J.D., Burns K.H. A map of mobile DNA insertions in the NCI-60 human cancer cell panel. Mob. DNA 7:20-20(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005 Li J., Zhao W., Akbani R., Liu W., Ju Z., Ling S., Vellano C.P., Roebuck P., Yu Q., Eterovic A.K., Byers L.A., Davies M.A., Deng W., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y., Mills G.B., Liang H. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell 31:225-239(2017)
PubMed=28601559; DOI=10.1016/j.cels.2017.05.009 Bekker-Jensen D.B., Kelstrup C.D., Batth T.S., Larsen S.C., Haldrup C., Bramsen J.B., Sorensen K.D., Hoyer S., Orntoft T.F., Andersen C.L., Nielsen M.L., Olsen J.V. An optimized shotgun strategy for the rapid generation of comprehensive human proteomes. Cell Syst. 4:587-599.e4(2017)
PubMed=28746345; DOI=10.1371/journal.pone.0181081 Sarin N., Engel F., Kalayda G.V., Mannewitz M., Cinatl J. Jr., Rothweiler F., Michaelis M., Saafan H., Ritter C.A., Jaehde U., Frotschl R. Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS ONE 12:E0181081-E0181081(2017)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051 Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F. Differential effector engagement by oncogenic KRAS. Cell Rep. 22:1889-1902(2018)
PubMed=29457907; DOI=10.1021/acs.jproteome.7b00782 Tomin T., Fritz K., Gindlhuber J., Waldherr L., Pucher B., Thallinger G.G., Nomura D.K., Schittmayer M., Birner-Gruenberger R. Deletion of adipose triglyceride lipase links triacylglycerol accumulation to a more-aggressive phenotype in A549 lung carcinoma cells. J. Proteome Res. 17:1415-1425(2018)
PubMed=29718670; DOI=10.1021/acs.jproteome.8b00165 Clark D.J., Hu Y., Bocik W., Chen L., Schnaubelt M., Roberts R., Shah P., Whiteley G., Zhang H. Evaluation of NCI-7 cell line panel as a reference material for clinical proteomics. J. Proteome Res. 17:2205-2215(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747 Dutil J., Chen Z., Monteiro A.N., Teer J.K., Eschrich S.A. An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines. Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3 Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J., Gelfand E.T., Bielski C.M., Li H., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569:503-508(2019) |
||
验收细胞注意事项
1、收到人肺癌细胞A549细胞,请查看瓶子是否有破裂,培养基是否漏出,是否浑浊,如有请尽快联系。
2、收到人肺癌细胞A549细胞,如包装完好,请在显微镜下观察细胞。,由于运输过程中的问题,细胞培养瓶中的贴壁细胞有可能从瓶壁中脱落下来,显微镜下观察会出现细胞悬浮的情况,出现此状态时,请不要打开细胞培养瓶,应立即将培养瓶置于细胞培养箱里静止 3-5 小时左右,让细胞先稳定下,再于显微镜下观察,此时多数细胞会重新贴附于瓶壁。如细胞仍不能贴壁,请用台盼蓝染色法鉴定细胞活力,如台盼蓝染色证实细胞活力正常请按悬浮细胞的方法处理。
3、收到人肺癌细胞A549细胞后,请镜下观察细胞,用恰当方式处理细胞。若悬浮的细胞较多,请离心收集细胞,接种到一个新的培养瓶中。弃掉原液,使用新鲜配制的培养基,使用进口胎牛血清。刚接到细胞,若细胞不多时 血清浓度可以加到 15%去培养。若细胞迏到 80%左右 ,血清浓度还是在 10%。
4、收到人肺癌细胞A549细胞时如无异常情况 ,请在显微镜下观察细胞密度,如为贴壁细胞,未超过80%汇合度时,将培养瓶中培养基吸出,留下 5-10ML 培养基继续培养:超过 80%汇合度时,请按细胞培养条件传代培养。如为悬浮细胞,吸出培养液,1000 转/分钟离心 3 分钟,吸出上清,管底细胞用新鲜培养基悬浮细胞后移回培养瓶。
5、将培养瓶置于 37℃培养箱中培养,盖子微微拧松。吸出的培养基可以保存在灭菌过的瓶子里,存放于 4℃冰箱,以备不时之需。
6、24 小时后,人肺癌细胞A549细胞形态已恢复并贴满瓶壁,即可传代。(贴壁细胞)将培养瓶里的培养基倒去,加 3-5ml(以能覆盖细胞生长面为准)PBS 或 Hanks’液洗涤后弃去。加 0.5-1ml 0.25%含 EDTA 的胰酶消化,消化时间以具体细胞为准,一般 1-3 分钟,不超过 5 分钟。可以放入37℃培养箱消化。轻轻晃动瓶壁,见细胞脱落下来,加入 3-5ml 培养基终止消化。用移液管轻轻吹打瓶壁上的细胞,使之完全脱落,然后将溶液吸入离心管内离心,1000rpm/5min。弃上清,视细胞数量决定分瓶数,一般一传二,如细胞量多可一传三,有些细胞不易传得过稀,有些生长较快的细胞则可以多传几瓶,以具体细胞和经验为准。(悬浮细胞)用移液管轻轻吹打瓶壁,直接将溶液吸入离心管离心即可。
7、贴壁细胞 ,悬浮细胞。严格无菌操作。换液时,换新的细胞培养瓶和换新鲜的培养液,37℃,5%CO2 培养。
特别提醒: 原瓶中培养基不宜继续使用,请更换新鲜培养基培养。