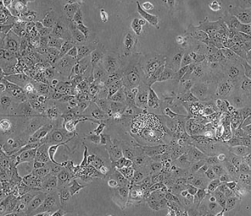
人非小细胞肺癌细胞HCC827
BLUEFBIO™ Product Sheet
|
细胞名称 |
人非小细胞肺癌细胞HCC827 |
|
|
|
货物编码 |
BFN60800668 |
||
|
产品规格 |
T25培养瓶x1 |
1.5ml冻存管x2 |
|
|
细胞数量 |
1x10^6 |
1x10^6 |
|
|
保存温度 |
37℃ |
-198℃ |
|
|
运输方式 |
常温保温运输 |
干冰运输 |
|
|
安全等级 |
1 |
||
|
用途限制 |
仅供科研用途 1类 |
||
|
培养体系 |
DMEM高糖培养基(Hyclone)+10%胎牛血清(Gibco)+1%双抗(Hyclone) |
||
|
培养温度 |
37℃ |
二氧化碳浓度 |
5% |
|
简介 |
人非小细胞肺癌细胞HCC827 细胞建于1994年三月。这株肺腺癌在EGFR酪氨酸激酶区域有一个获得性突变(E746-A750缺失)。患者在25岁到26岁时每个月抽1包烟。在诊断前12年不再抽烟。 |
||
|
注释 |
Part of: Cancer Cell Line Encyclopedia (CCLE) project. Part of: COSMIC cell lines project. Part of: MD Anderson Cell Lines Project. Doubling time: 71 hours (PubMed=25984343); 28 hours (ATCC); ~50-60 hours (DSMZ). Microsatellite instability: Stable (MSS) (Sanger). Omics: Deep exome analysis. Omics: Deep proteome analysis. Omics: Deep RNAseq analysis. Omics: DNA methylation analysis. Omics: Protein expression by reverse-phase protein arrays. Omics: shRNA library screening. Omics: SNP array analysis. Omics: Transcriptome analysis. |
||
|
STR信息 |
Amelogenin:X;CSF1PO:11;D13S317:9;D16S539:12;D18S51:13;D19S433:14;D21S11:31;D2S1338:17,24;D3S1358:17;D5S818:12;D7S820:11,12;D8S1179:12;FGA:22,24;TH01:6;TPOX:8;vWA:18 |
||
|
参考文献 |
PubMed=10987304 Girard L., Zochbauer-Muller S., Virmani A.K., Gazdar A.F., Minna J.D. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 60:4894-4906(2000)
PubMed=11314036; DOI=10.1038/sj.onc.1204211 Forgacs E., Wren J.D., Kamibayashi C., Kondo M., Xu X.L., Markowitz S.D., Tomlinson G.E., Muller C.Y., Gazdar A.F., Garner H.R., Minna J.D. Searching for microsatellite mutations in coding regions in lung, breast, ovarian and colorectal cancers. Oncogene 20:1005-1009(2001)
PubMed=18083107; DOI=10.1016/j.cell.2007.11.025 Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J.M., Yuan J., Bakalarski C.E., Villen J., Kornhauser J.M., Smith B., Li D., Zhou X., Gygi S.P., Gu T.-L., Polakiewicz R.D., Rush J., Comb M.J. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131:1190-1203(2007)
PubMed=20215515; DOI=10.1158/0008-5472.CAN-09-3458 Rothenberg S.M., Mohapatra G., Rivera M.N., Winokur D., Greninger P., Nitta M., Sadow P.M., Sooriyakumar G., Brannigan B.W., Ulman M.J., Perera R.M., Wang R., Tam A., Ma X.-J., Erlander M., Sgroi D.C., Rocco J.W., Lingen M.W., Cohen E.E.W., Louis D.N., Settleman J., Haber D.A. A genome-wide screen for microdeletions reveals disruption of polarity complex genes in diverse human cancers. Cancer Res. 70:2158-2164(2010)
PubMed=20679594; DOI=10.1093/jnci/djq279 Gazdar A.F., Girard L., Lockwood W.W., Lam W.L., Minna J.D. Lung cancer cell lines as tools for biomedical discovery and research. J. Natl. Cancer Inst. 102:1310-1321(2010)
PubMed=22460905; DOI=10.1038/nature11003 Barretina J.G., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.K., Yu J., Aspesi P. Jr., de Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., MacConaill L.E., Winckler W., Reich M., Li N., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483:603-607(2012)
PubMed=22961666; DOI=10.1158/2159-8290.CD-12-0112 Byers L.A., Wang J., Nilsson M.B., Fujimoto J., Saintigny P., Yordy J., Giri U., Peyton M., Fan Y.H., Diao L., Masrorpour F., Shen L., Liu W., Duchemann B., Tumula P., Bhardwaj V., Welsh J., Weber S., Glisson B.S., Kalhor N., Wistuba I.I., Girard L., Lippman S.M., Mills G.B., Coombes K.R., Weinstein J.N., Minna J.D., Heymach J.V. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2:798-811(2012)
PubMed=23733853; DOI=10.1101/gr.152322.112 Jia P., Jin H., Meador C.B., Xia J., Ohashi K., Liu L., Pirazzoli V., Dahlman K.B., Politi K., Michor F., Zhao Z., Pao W. Next-generation sequencing of paired tyrosine kinase inhibitor-sensitive and -resistant EGFR mutant lung cancer cell lines identifies spectrum of DNA changes associated with drug resistance. Genome Res. 23:1434-1445(2013)
PubMed=25960936; DOI=10.4161/21624011.2014.954893 Boegel S., Lower M., Bukur T., Sahin U., Castle J.C. A catalog of HLA type, HLA expression, and neo-epitope candidates in human cancer cell lines. OncoImmunology 3:E954893-E954893(2014)
PubMed=25984343; DOI=10.1038/sdata.2014.35 Cowley G.S., Weir B.A., Vazquez F., Tamayo P., Scott J.A., Rusin S., East-Seletsky A., Ali L.D., Gerath W.F.J., Pantel S.E., Lizotte P.H., Jiang G., Hsiao J., Tsherniak A., Dwinell E., Aoyama S., Okamoto M., Harrington W., Gelfand E., Green T.M., Tomko M.J., Gopal S., Wong T.C., Li H., Howell S., Stransky N., Liefeld T., Jang D., Bistline J., Hill Meyers B., Armstrong S.A., Anderson K.C., Stegmaier K., Reich M., Pellman D., Boehm J.S., Mesirov J.P., Golub T.R., Root D.E., Hahn W.C. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci. Data 1:140035-140035(2014)
PubMed=25485619; DOI=10.1038/nbt.3080 Klijn C., Durinck S., Stawiski E.W., Haverty P.M., Jiang Z., Liu H., Degenhardt J., Mayba O., Gnad F., Liu J., Pau G., Reeder J., Cao Y., Mukhyala K., Selvaraj S.K., Yu M., Zynda G.J., Brauer M.J., Wu T.D., Gentleman R.C., Manning G., Yauch R.L., Bourgon R., Stokoe D., Modrusan Z., Neve R.M., de Sauvage F.J., Settleman J., Seshagiri S., Zhang Z. A comprehensive transcriptional portrait of human cancer cell lines. Nat. Biotechnol. 33:306-312(2015)
PubMed=26361996; DOI=10.1016/j.jprot.2015.09.003 Grundner-Culemann K., Dybowski J.N., Klammer M., Tebbe A., Schaab C., Daub H. Comparative proteome analysis across non-small cell lung cancer cell lines. J. Proteomics 130:1-10(2016)
PubMed=27397505; DOI=10.1016/j.cell.2016.06.017 Iorio F., Knijnenburg T.A., Vis D.J., Bignell G.R., Menden M.P., Schubert M., Aben N., Goncalves E., Barthorpe S., Lightfoot H., Cokelaer T., Greninger P., van Dyk E., Chang H., de Silva H., Heyn H., Deng X., Egan R.K., Liu Q., Mironenko T., Mitropoulos X., Richardson L., Wang J., Zhang T., Moran S., Sayols S., Soleimani M., Tamborero D., Lopez-Bigas N., Ross-Macdonald P., Esteller M., Gray N.S., Haber D.A., Stratton M.R., Benes C.H., Wessels L.F.A., Saez-Rodriguez J., McDermott U., Garnett M.J. A landscape of pharmacogenomic interactions in cancer. Cell 166:740-754(2016)
PubMed=28196595; DOI=10.1016/j.ccell.2017.01.005 Li J., Zhao W., Akbani R., Liu W., Ju Z., Ling S., Vellano C.P., Roebuck P., Yu Q., Eterovic A.K., Byers L.A., Davies M.A., Deng W., Gopal Y.N.V., Chen G., von Euw E.M., Slamon D.J., Conklin D., Heymach J.V., Gazdar A.F., Minna J.D., Myers J.N., Lu Y., Mills G.B., Liang H. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell 31:225-239(2017)
PubMed=29444439; DOI=10.1016/j.celrep.2018.01.051 Yuan T.L., Amzallag A., Bagni R., Yi M., Afghani S., Burgan W., Fer N., Strathern L.A., Powell K., Smith B., Waters A.M., Drubin D., Thomson T., Liao R., Greninger P., Stein G.T., Murchie E., Cortez E., Egan R.K., Procter L., Bess M., Cheng K.T., Lee C.-S., Lee L.C., Fellmann C., Stephens R., Luo J., Lowe S.W., Benes C.H., McCormick F. Differential effector engagement by oncogenic KRAS. Cell Rep. 22:1889-1902(2018)
PubMed=30894373; DOI=10.1158/0008-5472.CAN-18-2747 Dutil J., Chen Z., Monteiro A.N., Teer J.K., Eschrich S.A. An interactive resource to probe genetic diversity and estimated ancestry in cancer cell lines. Cancer Res. 79:1263-1273(2019)
PubMed=31068700; DOI=10.1038/s41586-019-1186-3 Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J., Gelfand E.T., Bielski C.M., Li H., Hu K., Andreev-Drakhlin A.Y., Kim J., Hess J.M., Haas B.J., Aguet F., Weir B.A., Rothberg M.V., Paolella B.R., Lawrence M.S., Akbani R., Lu Y., Tiv H.L., Gokhale P.C., de Weck A., Mansour A.A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J.M., Porter D.A., Jones M.D., Golji J., Caponigro G., Taylor J.E., Dunning C.M., Creech A.L., Warren A.C., McFarland J.M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y.E., Cherniack A.D., Tsherniak A., Vazquez F., Jaffe J.D., Lane A.A., Weinstock D.M., Johannessen C.M., Morrissey M.P., Stegmeier F., Schlegel R., Hahn W.C., Getz G., Mills G.B., Boehm J.S., Golub T.R., Garraway L.A., Sellers W.R. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569:503-508(2019) |
||
验收细胞注意事项
1、收到人非小细胞肺癌细胞HCC827细胞,请查看瓶子是否有破裂,培养基是否漏出,是否浑浊,如有请尽快联系。
2、收到人非小细胞肺癌细胞HCC827细胞,如包装完好,请在显微镜下观察细胞。,由于运输过程中的问题,细胞培养瓶中的贴壁细胞有可能从瓶壁中脱落下来,显微镜下观察会出现细胞悬浮的情况,出现此状态时,请不要打开细胞培养瓶,应立即将培养瓶置于细胞培养箱里静止 3-5 小时左右,让细胞先稳定下,再于显微镜下观察,此时多数细胞会重新贴附于瓶壁。如细胞仍不能贴壁,请用台盼蓝染色法鉴定细胞活力,如台盼蓝染色证实细胞活力正常请按悬浮细胞的方法处理。
3、收到人非小细胞肺癌细胞HCC827细胞后,请镜下观察细胞,用恰当方式处理细胞。若悬浮的细胞较多,请离心收集细胞,接种到一个新的培养瓶中。弃掉原液,使用新鲜配制的培养基,使用进口胎牛血清。刚接到细胞,若细胞不多时 血清浓度可以加到 15%去培养。若细胞迏到 80%左右 ,血清浓度还是在 10%。
4、收到人非小细胞肺癌细胞HCC827细胞时如无异常情况 ,请在显微镜下观察细胞密度,如为贴壁细胞,未超过80%汇合度时,将培养瓶中培养基吸出,留下 5-10ML 培养基继续培养:超过 80%汇合度时,请按细胞培养条件传代培养。如为悬浮细胞,吸出培养液,1000 转/分钟离心 3 分钟,吸出上清,管底细胞用新鲜培养基悬浮细胞后移回培养瓶。
5、将培养瓶置于 37℃培养箱中培养,盖子微微拧松。吸出的培养基可以保存在灭菌过的瓶子里,存放于 4℃冰箱,以备不时之需。
6、24 小时后,人非小细胞肺癌细胞HCC827细胞形态已恢复并贴满瓶壁,即可传代。(贴壁细胞)将培养瓶里的培养基倒去,加 3-5ml(以能覆盖细胞生长面为准)PBS 或 Hanks’液洗涤后弃去。加 0.5-1ml 0.25%含 EDTA 的胰酶消化,消化时间以具体细胞为准,一般 1-3 分钟,不超过 5 分钟。可以放入37℃培养箱消化。轻轻晃动瓶壁,见细胞脱落下来,加入 3-5ml 培养基终止消化。用移液管轻轻吹打瓶壁上的细胞,使之完全脱落,然后将溶液吸入离心管内离心,1000rpm/5min。弃上清,视细胞数量决定分瓶数,一般一传二,如细胞量多可一传三,有些细胞不易传得过稀,有些生长较快的细胞则可以多传几瓶,以具体细胞和经验为准。(悬浮细胞)用移液管轻轻吹打瓶壁,直接将溶液吸入离心管离心即可。
7、贴壁细胞 ,悬浮细胞。严格无菌操作。换液时,换新的细胞培养瓶和换新鲜的培养液,37℃,5%CO2 培养。
特别提醒: 原瓶中培养基不宜继续使用,请更换新鲜培养基培养。